
GigaTime
GigaTIME is a multimodal AI model for translating routinely available hematoxylin and eosin (H&E) pathology slides to virtual multiplex immunofluorescence (mIF) images.
Developed in collaboration with Providence and the University of Washington, GigaTIME was trained on a Providence dataset of 40 million cells with paired H&E and mIF images across 21 protein channels.
Digital pathology transforms a microscopy slide of stained tumor tissue into a high-resolution digital image, revealing details of cell morphology such as nucleus and cytoplasm. Such a slide only costs $5 to $10 per image and has become routinely available in cancer care. It is well known that H&E-based cell morphology contains information about the cellular states. However, prior works are generally limited to average biomarker status across the entire tissue. GigaTIME thus represents a major step forward by learning to predict spatially resolved, single-cell states essential for tumor microenvironment modeling. In turn, this enables us to generate a virtual population of mIF images for large-scale TIME analysis (Figure 1).
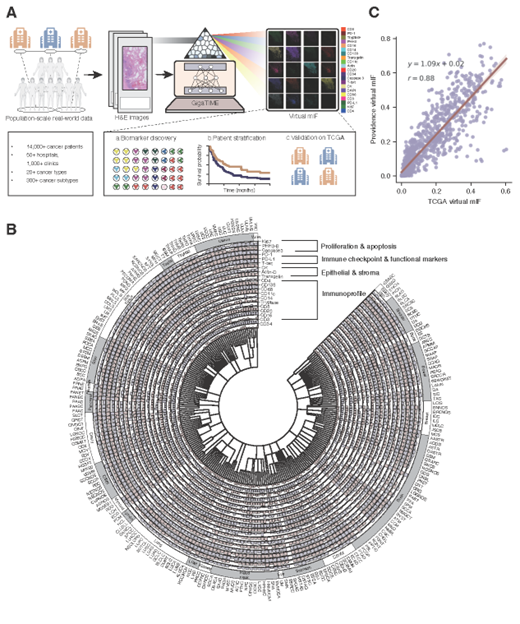
Figure 1: GigaTIME enables population-scale tumor immune microenvironment (TIME) analysis. A, GigaTIME inputs a hematoxylin and eosin (H&E) whole-slide image and outputs multiplex immunofluorescence (mIF) across 21 protein channels. By applying GigaTIME to 14,256 patients, we generated a virtual population with mIF information, leading to population-scale discovery on clinical biomarkers and patient stratification, with independent validation on TCGA. B, Circular plot visualizing a TIME spectrum encompassing the GigaTIME-translated virtual mIF activation scores across different protein channels at the population scale, where each channel is represented as an individual circular bar chart segment. The inner circle encodes OncoTree, which classifies 14,256 patients into 306 subtypes across 24 cancer types. The outer circle groups these activations by cancer types, allowing visual comparison across major categories. C, Scatter plot comparing the subtype-level GigaTIME-translated virtual mIF activations between TCGA and Providence virtual populations. Each dot denotes the average activation score of a protein channel among all tumors of a cancer subtype..
Researchers applied GigaTIME to 14,256 cancer patients from 51 hospitals and over a thousand clinics within the Providence system. This effort generated a virtual population of around 300,000 mIF images spanning 24 cancer types and 306 cancer subtypes. This virtual population uncovered 1,234 statistically significant associations linking mIF protein activations with key clinical attributes such as biomarkers, staging, and patient survival. Independent external validation on 10,200 Cancer Genome Atlas (TCGA) patients further corroborated our findings.
To the researchers’ knowledge, this is the first population-scale study of tumor immune microenvironment (TIME) based on spatial proteomics. Such studies were previously infeasible due to the scarcity of mIF data. By translating readily available H&E pathology slides into high-resolution virtual mIF data, GigaTIME provides a novel research framework for exploring precision immuno-oncology through population-scale TIME analysis and discovery. The GigaTIME model is publicly available to help accelerate clinical research in precision oncology.